Convert Freesurfer Outputs and Calculate Dist Q
Written by Owen Phillips. Email Dr. Katherine Narr if you have any questions. For more information, see the protocols page.
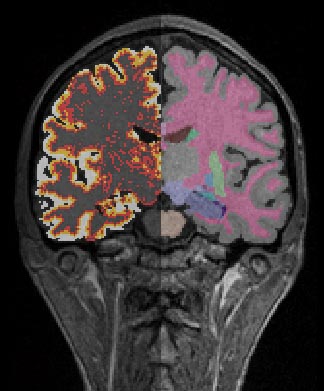
Dist Q and Aseg from Conversion Pipeline
Once Freesurfer has finished running, you can run the pipeline Freesurfer_masking_and_convertion_RAS. This will take the .mgz outputs from Freesurfer and convert them to analyze format. Furthermore, it will combine the segmented labels to generate a useful brainmask. This brainmask is then used to generate hemisphere masks, which are in turned used to generate tissue segmented images (PVC) and finally thickness distance quick (Dist Q) images.
- Required inputs: The inputs will be found in your freesurfer directory mri folder. For example: */schizo/ANATOMY_CLASS/FS/M203_INSTRUCTOR_27/mri
- T1.mgz
- aseg.mgz
- ribbon.mgz
- brainmask.mgz
- Outputs:
- T1.img
- T1 Masked.img
- Dist Q Left Hemisphere (distance quick) (analyze format .img/.hdr)
- Dist Q Left Hemisphere (distance quick) (Minc format .mnc)
- Tissue Classified (PVC) Left Hemisphere
- Left Hemisphere mask
- White Matter Left Hemisphere
- White Matter Right Hemisphere
- Tissue Classified (PVC) Right Hemisphere
- Right Hemisphere mask
- Dist Q Right Hemisphere (distance quick) (analyze format .img/.hdr)
- Dist Q Right Hemisphere (distance quick) (Minc format .mnc)
- Aseg (Freesurfer generated labels .img)
- Ribbon (Freesurfere generated image surface file .img)
- Freesurfer's automatic brainmask (.img) - This file is useful as backup but is otherwise not used.
- T1 automasked (.img) - This file is the T1 image that has been masked by the freesurfer automatic brainmask.
- After running, check to insure you have to correct left right orientation. This can change depending on your data set. If you need to fix it, you can adjust the MRI Convert module from RAS orientation to LAS orientation.


