Written by Owen Phillips. Email Dr. Katherine Narr if you have any questions. For more information, see the protocols page.
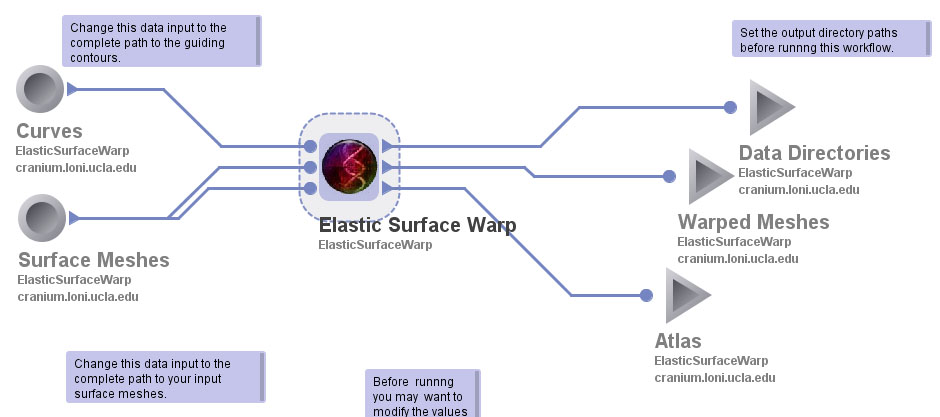
After Sulcal Lines are combined to one file, the data is ready to be run through the warping pipeline. The purpose of the warping pipeline is to use the sulcal linemarks to drive homology between they subjects surfaces. You can find a more comprehensive overview here: http://www.loni.ucla.edu/twiki/bin/view/MAST/ElasticSurfaceWarpProtocolDataAssumptions?skin=plain. To run the pipeline you will need to the LONI Pipeline as well as the ElasticSurfaceWarp.pipe (this can be downloaded by right clicking).
It is important to decide the kind of analysis you want to do before warping. Unless you are doing an asymmetry analysis, the left and right hemispheres should be run as separate pipelines. This will preserve their original left and right orientation. If they are run together, both hemispheres will be flipped into the left hemisphere space.
Set the inputs:
Set the ouputs: