You will need the following:
You will be working with the following scripts (which can be found and/or copied from /cxfs/schizo/family_study/scripts/medial_analysis, and whose execution will be described below):
If your lines are objects are already colored and all of your lines are definitely in the correct directions and named correctly, you can skip these steps. However, making sure that all of your lines look good before you run any of the scripts will end up saving you a lot of time.
Here we list, one by one, each of the scripts that you will need to run and/or modify to make your flatmaps. Again, these are located at /cxfs/schizo/family_study/scripts/medial_analysis/.
yourname@inire% cd /cxfs/schizo/family_study/scripts/medial_analysis/.
This script does exactly what it says. It copies lines to another directory, and flips the right hemisphere lines into left space. You will need to modify the script for your data as follows:
Modifications:
Run this script on inire. The directory shouldn't matter, unless you haven't given a full path to your list file, and then you should be in the list file's directory.
yourname@inire% ./01_copy_flip_lines.csh
This script divides your central sulcus (line 1), transverse occipital sulcus (line 21), and superior frontal sulcus (line 5) into two segments. These lines can cause problems with the warping if they remain as a whole line. It also converts your lines from .obj files to UCFs.
Modifications
This script is very simple. It just moves all of the redigitized files to the subdirectories for averaging.
Modifications
This script computes average and variance maps for the lines in the RSPs directory. These averages will be used in the warping.
For this script, you shouldn't have to modify anything, but you will have to specify each one of your subject directories as an argument. You can either do this in a loop on the command line [see (1) and (2)] or by specifying each in one line [see (3)].
This script creates the Left_CORT and Right_CORT analysis directories in your subject directories, and flips your right hemisphere objects into left space. The left and right hemisphere objects are then copied to the Left_CORT and Right_CORT directories, respectively.
Modifications
This script shortens all of your ucfs from the MED_RSPs directory within each subject directory and copies them to the Left_CORT or Right_CORT directory as SHORTRSP3_*. Again, change the directory at line 7 to your own directory.
This script will create flattened lines (FLATs and DUALFLATs) for use in the warping. Lines that "fall off" the brain during warping will be recorded in the FALL_OFF directories (but note that these are not true ucfs, just a list of the numbers). Before running this script, be sure you have the following in your Left_CORT or Right_CORT directory:
Modify all lines containing "family_study" to reflect directories appropriate to your study.
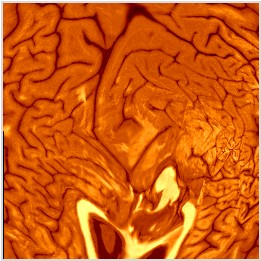
To view the resulting flat map (pre-warping), load DUALflat.uvl in seg. Your flat map will look something like the case below, for RsurrL:
This will copy the flattened lines into a directory to make the average target/template that is used to warp the flat maps. Two templates are made, one from FLATSHOTRSP3s and one from DUALFLATSHORTRSP3s.
Modify lines containing "family_study" with your directory names.
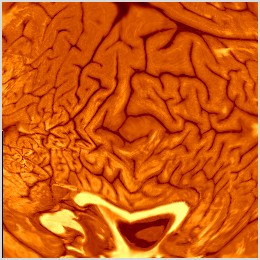
Original RsurrL flat map and its corresponding warp:
Place your mouse over the original flat map on the left to see the change induced by warping and toggle between the unwarped and warped versions (on the right).


Bad warping of RsurrL flat map and its color map:


After this script, you should end up with DUALflat warps and flat warps. Mouse over the LsurrR (flatmaps, on left) and RsurrL (dualflats, on right) images below to see the resulting warps. In the screenshots below from seg, both images represent the left hemisphere.

This script converts the BIG color maps (UIFs) to whole brain 3D UCFs. All of the L hems get blended into a UCF in the *L.csh script, and all of the R hems are blended into a UCF in the *R.csh script.
You will be blending prEw_LsurrR_BIGord.uif, DECprEw_LsurrR_BIGord.uif, prEw_RsurrL_BIGord.uif, and DECprEw_RsurrL_BIGord.uif into ${subj}_prEw_BIG_whole_Lhem_blend.ucf
You will use this resulting UCF file in your statistical analyses.
Creates a VAR and AVG map from the left hemisphere inflated hemisphere from the previous step.