Skull Stripping in MultiTracer Using the TraceWalker Utility
Software Tool Developed by Roger Woods, protocol written by Owen Phillips, August 2007. Email Dr. Katherine Narr with questions or comments.
This protocol is for skull stripping a brain while leaving out the cerebellum.
- If you do not have Multitracer.jar you can download it by clicking on the link.
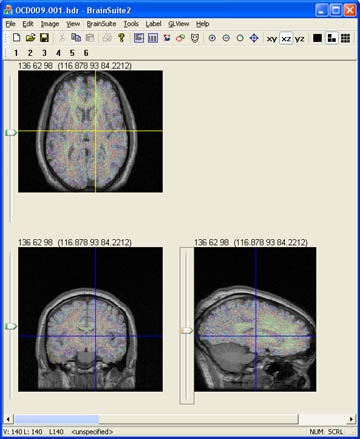
- Open the brain volume of interest in MultiTracer. Tracing should be done on the axial view. You can increase the magnification of the image by entering a value of 1-8 in the Magnify by: box. For the coronal and sagittal views the image can be flipped vertically and magnified under the Orient tools.

- In order for the skull stripping to work you must designate two types of contours. The first contour must be your brain contour. For this tutorial the first contour will be called Brain and I will set it to a yellow color. The second contour will be called Non_Brain and it will be set to a red color. The naming system is arbitrary but the order in which you set the contours is critical. The skull stripping tool assumes the first contour in your list is Brain and it will fail if this order is not followed.
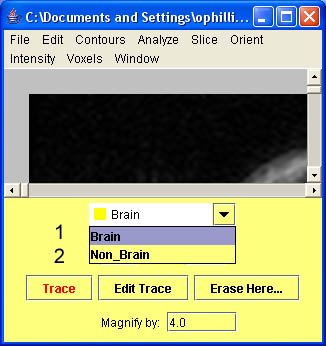
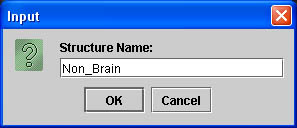
- To add a structure click on Contours and scroll down to Add contour. A box will pop up and you will be able to name your new structure.
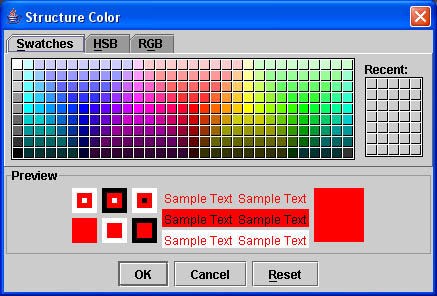
- To change the color of a contour Click Contour and scroll down to Structure Color. You can easily set the color of your structure to whatever you prefer. This step is not critical but it makes drawing the contours less confusing.
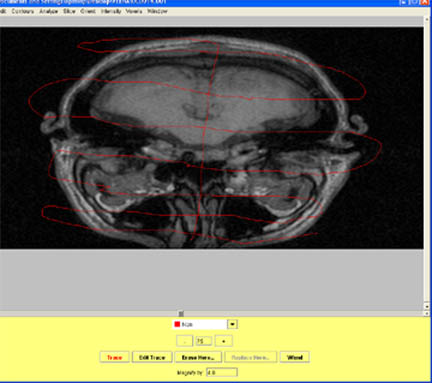
- Begin by drawing your contours inferior to the start of the temporal or occipital lobe on the axial view. Mark all structures before the brain starts as Non_Brain.
- Depending on the brain the Temporal or the occipital lobe will appear first. Once brain tissue appears mark it as brain. Mark all other structures as Non_Brain. The area around the eye orbits can be tricky to distinguish between brain and non brain. For these areas it is best to mark carefully around the brain as Non_Brain in order to cover all the intensity values. Also be sure to consult the coronal view as a reference. This region often produces the most problems with the final product. By marking every 5 slices and carefully marking all Brain and Non_Brain intensity values it will produce a better result.

- Watch for the Temporal and Occipital lobes to appear in the view together. In some brains the frontal lobe may appear earlier. In either case continue to mark Brain and Non_Brain.

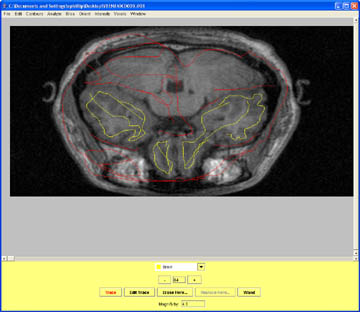
- Once the brainstem connects with the brain in the medial temporal region include the brainstem. Watch for the optic nerves and mark these as non brain. They are located just below the brainstem and they are bright white, typical of nerve tracts.

- Once the Corpus Callosum appears and connects the right and left hemispheres mark the CSF anterior to the Corpus Callosum as Brain and the CSF posterior as Non_Brain.
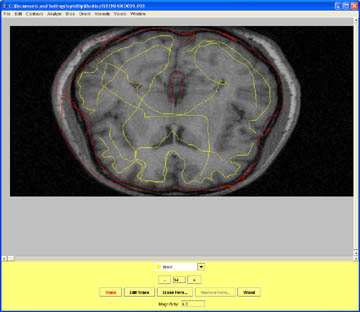
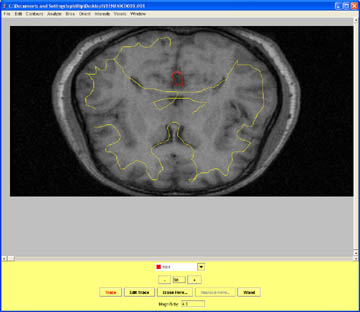
- The superior aspects of the brain can cause trouble if the meninges are not carefully labeled as Non_Brain.
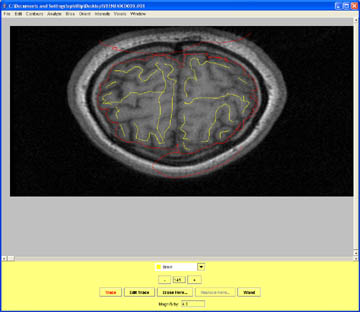
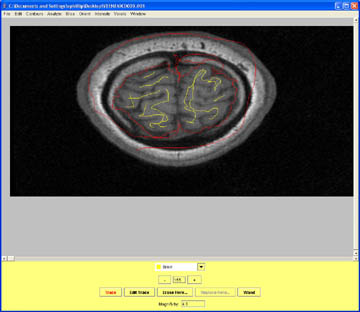
- Mark the slice after the most superior aspect of the brain as Non_Brain and save your contour data.
- A real world example using the Terminal:
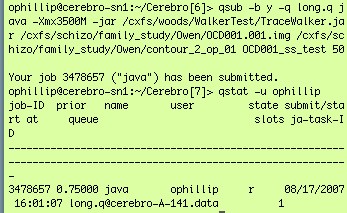
- To run the script using the command line, first connect to qsub.loni.ucla.edu, then run the following command:
qsub -b y -q long.q java -Xmx3500M -jar /dir/TraceWalker.jar /(dir)/(image_file) /(dir)/(contour) /OUTPUT_DIR/(output_file) (Threshold value)
I have found that thresholds values between the range of 30-50 work best. But you may have to run your data multiple times to find the threshold value that works best for you.
- You can also run locally by using the terminal on your computer. Download the TraceWalker.jar file first.
- Sample Command line in the terminal: Java -jar -Xmx1500M TraceWalker.jar in.img contour_file out.img 30
If you get an error saying "could not reserve enough space for object heap, try giving the command more memory, such as -Xmx2000M.
- You can also run your processing using the LONI Pipeline. You can download the TraceWalker.pipe module for use in the pipeline by following the link.

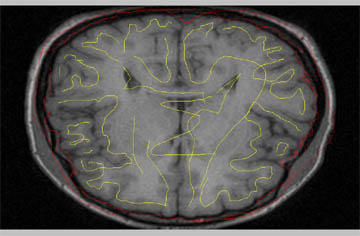
- Your skull stripped brain should look something like this. To learn how to convert this to a mask, visit the protocols page for coverting analyze files to masks in BrainSuite.
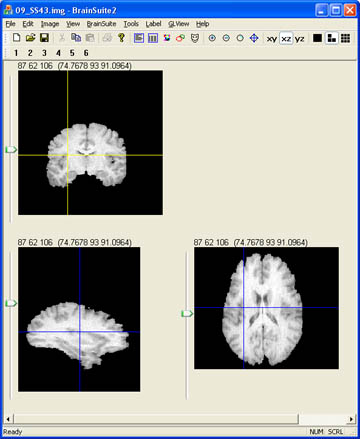
Some Useful tips:
A nice way to check how well your contours worked is to view the data in Brainsuite. If you are not familar with BrainSuite you can find more turorial information and downloads at brainsuite.usc.edu, or you can check out a few tutorials I have made under the Miscellaneous section of our protocols page.
- Load the original brain as your volume.
- Load your skull stripped brain as your Label.
- Check over the brain to insure you haven't cut out any brain tissue or that you still have meninges.
Notes: Some brains will require more contours then others depending on the areas of contrast. Occasionally the ventricles can be left out but this can be fixed by adding more contours or by changing the threshold value.
Common Questions:
- Q: How often should I mark contours?
A: I have found that if you want the best possible result, marking once every five slices will provide a great result. You can get by with fewer contours (1/10 or 1/15) in the middle of the brain but this often results in meninges finding their way onto your final product.
- Q: What should my contours look like?
A: It seems that everyone who tries using this utility comes up with their own way of drawing contours. What works best for me is first drawing a careful outline around the brain while watching for patches of meninges and including those as Non_Brain. Then I mark the skull and all other external structures as Non_Brain. I then carefully draw on the inside edge of the brain to give me a nice outline of the areas I want to include. Finally I add a few contours through the middle of my brain area to cover all possible intensity values. This is probably a bit excessive and I would welcome feedback from anyone who comes up with an improved way of marking contours. But this method produces a nice result.
Email: Dr. Katherine Narr if you have any questions or comments regarding this protocol.


















